Registration form and contact
Due to overwhelming interest, we have reached our full capacity for attendees. We are sorry to announce that registration is now closed.
You can ask your questions by mail using this email address : chemoinfo-school@unistra.fr
Abstract Submission
Abstracts should be submitted to the Organizers by e-mail before April 15th 2024.
The attachment should be in RTF or DOC format.
e-mail : chemoinfo-school@unistra.fr
The posters should be of the following format : A0, Portrait orientation.
The abstract should be prepared according to a set of rules :
You can download the following word document to use as your abstract template.
Abstract_Template
Download abstract template
Here is an abstract example :
Communication Title (Arial, 14 pts, bold, centered)
FirstName LastName1, FirstNme LastNme1, FirstNme LastNme 2,… (Arial, 11 pts, centered)
1Laboratory1, University/Institute/Society…, Address, postal code, City, Country.
2Laboratory2, … (Arial, italics, 11 pts, aligned left)
Abstract Text : Arial, simple line spacing, 11 pts, justified
A black&white figure is authorized, referenced in the text as : (figure 1).
Figure legend in Arial, 8 pts, simple line spacing.
Bibliography :
[1] X.X ; Y.Y. J. Catal. 163 (2006) 192-156. (Arial, 10 pts, aligned left)
Please note that the abstract must not go beyond an A4 page (21 x 29,7 cm).
Margins are fixed to 2cm.
Conference fees
Early registration (before April 30th 2024)
| Industrial |
1000 € |
| Academics |
700 € |
| Students |
500 € |
| Accompagnying person |
300 € |
| Pre-conference session : |
250 € |
After April 30th 2024
| Industrial |
1100 € |
| Academics |
750 € |
| Students |
550 € |
| Accompagnying person |
350 € |
| Pre-conference session : |
250 € |
The registration fees covers :
Conference sessions and materials,
Conference dinner on 27 June,
Welcome party,
Coffee breaks,
Evening « Beer and Bretzel » parties on 26 and 27 June
Lunches on 25, 26 and 27 June
Cultural program on 27 June
Location
Strasbourg Summer School - Strasbourg, France, 24 – 28 June 2024
Welcome to Strasbourg !
Its magnificent Cathedral, its fine food, "La Petite France" the typical quarter beloved of the tourists, not to mention the Grande-Ile, a UNESCO World Heritage site, have given it its reputation as a city anchored in History and Tradition.
Located in the "Esplanade" district, the Central Campus is near Strasbourg historical center. Also called "Esplanade Campus", it is the main University of Strasbourg Campus, regrouping University headquarters and many University of Strasbourg elements (including the "Tour de Chimie"), as well as the main part of the research units. Several independant high schools are present as well (Ecole Supérieure des Arts Décoratifs de Strasbourg and INSA).
Conference venue
The conference will be held at the European Doctoral College building in the central campus of the University of Strasbourg (46 Boulevard de la Victoire, 67000 Strasbourg).
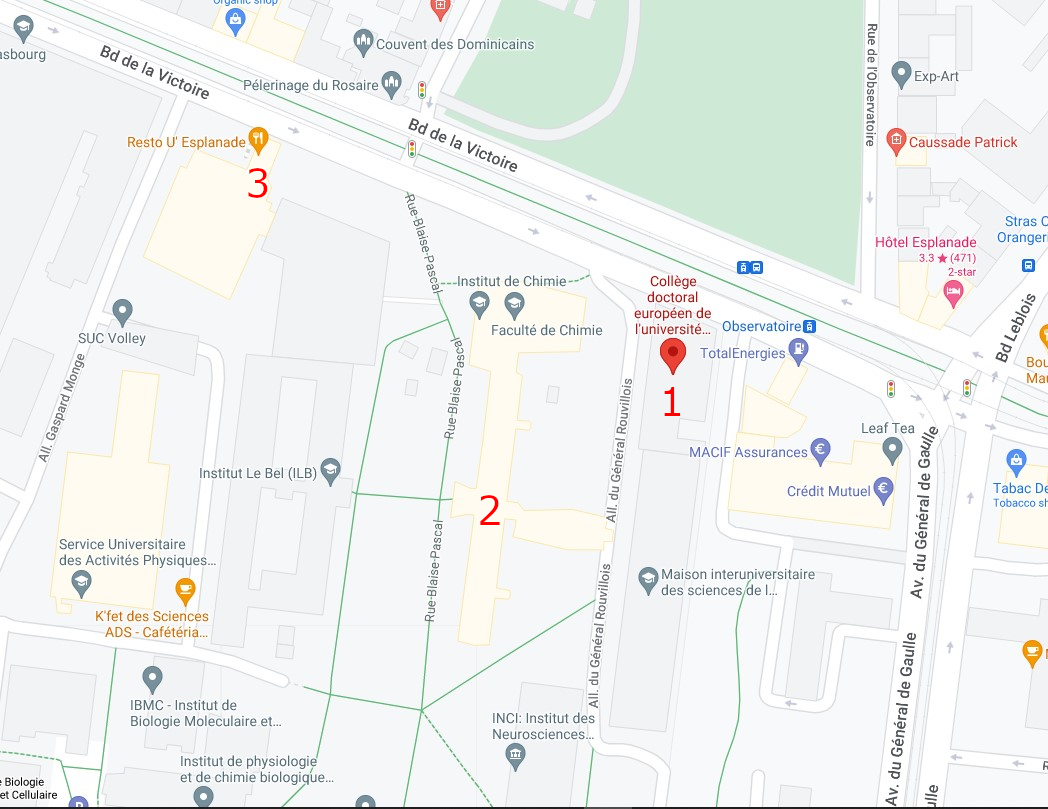
Campus map
1 : European Doctoral College (46 Bd de la Victoire, 67000 Strasbourg)
2 : Chemistry Tower (1 Rue Blaise Pascal, 67008 Strasbourg)
3 : Restaurant "le 32" (Boulevard de la Victoire, Strasbourg)
How to get to the Conference Venue ?
From the airport
- Take the shuttle train until the terminus Strasbourg Railway Station (see here)
- Take the tram line C, direction "Neuhof Rodolphe Reuss" and get off at the stop "Observatoire".
From the train station
Take the tram line C, direction "Neuhof / Rodolphe Reuss" and get off at the stop "Observatoire".
From a bus or tram station
The Central Campus is situated near the tram station "Observatoire" on lines C and E.
Please check the map of the city network Here
You can buy tram tickets via the CTS application (Google Play, App Store), or at each tram station.
By car
Coming from the North, on highway A4:
- Follow the directions to "Kehl".
- After the tunnel, stay on the left and follow the direction "Esplanade".
- Turn left and cross the Pont du Danube.
- Go straight on Rue d’Ankara and on Rue de Leicester.
- Turn left to rue de Londres.
- At traffic light, go straight across avenue du General de Gaulle and the tram rails.
- Enter the Place d’Athènes, which is in front of the Campus Central.
Coming from the South, on highway A35:
- Take the Exit "Place de l’Etoile-Neudorf", stay on the left lane.
- Follow the direction to Kehl.
- Continue as described above (directions for coming from the North on highway A4).
Important information !
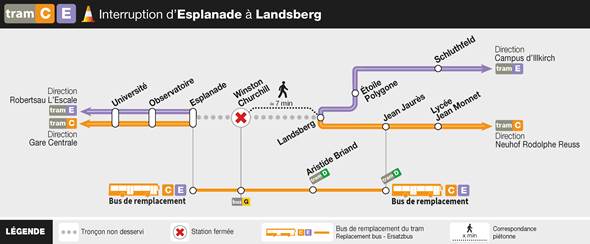
We would like to inform you that during the period from Monday, June 24th to Friday, August 23rd, 2024, inclusive, the tram company will be carrying out maintenance work on the tramway tracks. This will result in a complete interruption of service on the affected sections.
To ensure continuity of service, replacement buses will be put in place on the sections not served by the tramway during the same hours of operation.
Trams C and E are not running between Esplanade and Landsberg stations.
The northbound Tram C runs only between Gare Centrale and Esplanade.
The northbound Tram E runs only between Robertsau L’Escale and Esplanade.
The southbound Trams C and E are exceptionally running only between Campus d’Illkirch and Neuhof Rodolphe Reuss (with a route change at Landsberg).
Tram D is operating normally.
The map of the central campus is available here.
Photos by Jonathan Martz and Geneviève Engel.