We are happy to announce the 8th Strasbourg Summer School in Chemoinformatics (CS3-2022) to be held in Strasbourg, France, from 27th June to 1st July 2022.
CS3-2022 is a unique school mixing students, young and experienced scientists.
Registration is closed !
The program includes plenary lectures, poster session, oral presentations and hands-on tutorials. It covers the following topics:
· Artificial Intelligence in chemistry:
- Artificial Intelligence in chemistry
- Big Data analysis and visualization
- Chemical Reactions mining
- New trends in QSAR modeling
- Materials informatics
- In silico pharmacology
Among invited speakers, it is our pleasure to acknowledge the participation of many world-class scientists:
- J. Bajorath (Univ. Bonn, Germany)
- T. Langer (Univ. Vienna, Austria)
- G. Schneider (ETH, Zürich, Switzerland)
- A. Cherkasov (Univ. British Columbia, Canada)
- A. Tropsha (Univ. North Carolina, Chappel Hill, USA)
- P. Polishchuk (Palacký University Olomouc, Czech Republic)
- M. Rarey (Univ. Hamburg, Germany)
- O. Taboureau (Univ. Paris, France)
- J. Medina-Franco (Univ. Mexico)
- J. Kirchmair (Univ. Vienna, Austria)
- E. Benfenati (Ist. Ricerche Farm. Mario Negri, Milan, Italy)
- H. Senderowitz (Univ. Bar Ilan, Israel)
- C. Coley (MTI, USA)
- N. Stiefl (Novartis, Switzerland)
- O. Engkvist (ASTRAZENECA).
A pre-conference ”Crash course in Chemoinformatics” for the beginners will be organized on June 27th from 9:00 to 16:00 (for registered participants only).
A pre-conference "Interactive Workshop with SeeSAR" will be organized on June 27th from 14:00 to 16:00.
Please note that the CS3 program also includes several short oral communications (20 min) in order to enable software editors to present their latest developments used in particular scientific projects. The number of communications is limited, a “first come, first served” policy will be applied.
Tiny URL of this page: https://tinyurl.com/CS3-2022
QR code:
Informations about Strasbourg University
Location
Strasbourg Summer School - Strasbourg, France, 29 June - 3 July 2020
Welcome to Strasbourg !
Its magnificent Cathedral, its fine food, "La Petite France" the typical quarter beloved of the tourists, not to mention the Grande-Ile, a UNESCO World Heritage site, have given it its reputation as a city anchored in History and Tradition.
Located in the "Esplanade" district, the Central Campus is near Strasbourg historical center. Also called "Esplanade Campus", it is the main University of Strasbourg Campus, regrouping University headquarters and many University of Strasbourg elements (including the "Tour de Chimie"), as well as the main part of the research units. Several independant high schools are present as well (Ecole Supérieure des Arts Décoratifs de Strasbourg and INSA).
Conference venue
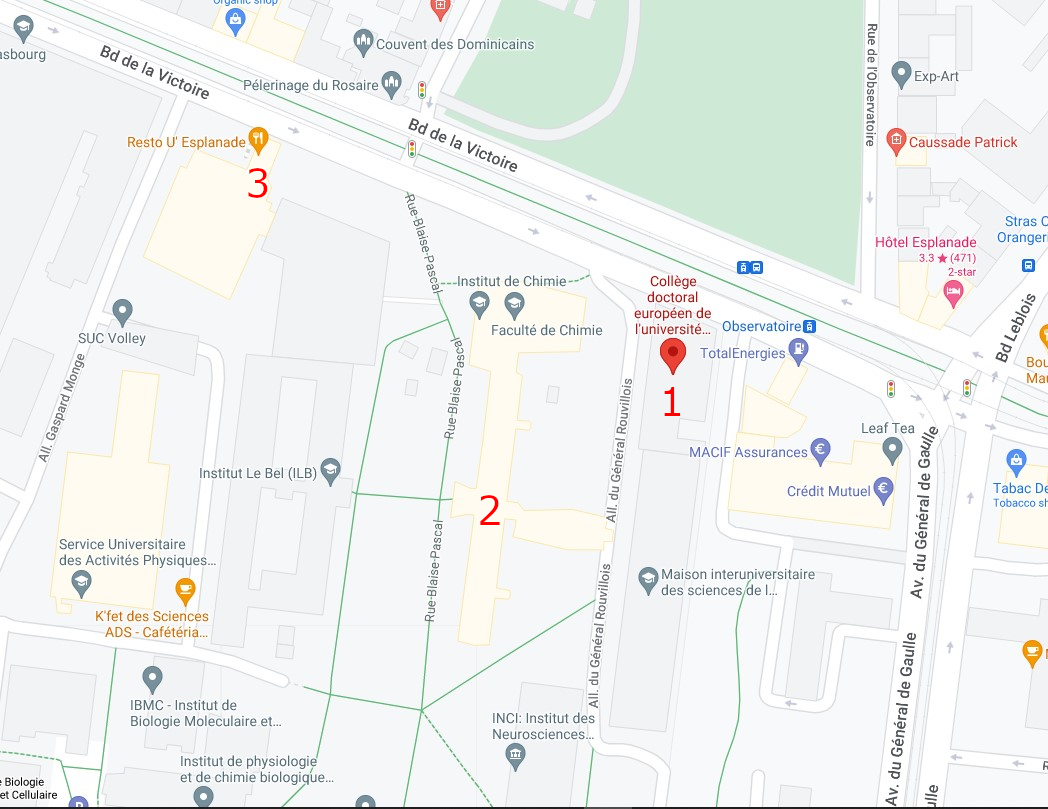
The conference will be held at the European Doctoral College building in the central campus of the University of Strasbourg (46 Boulevard de la Victoire, 67000 Strasbourg).
Campus map
1 : European Doctoral College (46 Bd de la Victoire, 67000 Strasbourg)
2 : Chemistry Tower (1 Rue Blaise Pascal, 67008 Strasbourg)
3 : Restaurant "le 32" (Boulevard de la Victoire, Strasbourg)
Social program map
Here are 3 proposed routes to get to the European Council (Av. de l’Europe, 67000 Strasbourg)
Route 1 : replacement bus + tram E
Route 2 : walking + bus 72 or L6
Route 3 : walking
Conference dinner map
1 : European Doctoral College (46 Bd de la Victoire, 67000 Strasbourg)
2 : Restaurant "la Bourse" (Place du Maréchal de Lattre de Tassigny, Strasbourg)
How to get to the Conference Venue ?
From the airport
- Take the shuttle train until the terminus Strasbourg Railway Station (see here)
- Take the tram line C, direction "Neuhof Rodolphe Reuss" and get off at the stop "Observatoire".
From the train station
Take the tram line C, direction "Neuhof / Rodolphe Reuss" and get off at the stop "Observatoire".
From a bus or tram station
The Central Campus is situated near the tram station "Observatoire" on lines C and E.
Please check the map of the city network Here
By car
Coming from the North, on highway A4:
- Follow the directions to "Kehl".
- After the tunnel, stay on the left and follow the direction "Esplanade".
- Turn left and cross the Pont du Danube.
- Go straight on Rue d’Ankara and on Rue de Leicester.
- Turn left to rue de Londres.
- At traffic light, go straight across avenue du General de Gaulle and the tram rails.
- Enter the Place d’Athènes, which is in front of the Campus Central.
Coming from the South, on highway A35:
- Take the Exit "Place de l’Etoile-Neudorf", stay on the left lane.
- Follow the direction to Kehl.
- Continue as described above (directions for coming from the North on highway A4).
Important information !
There will be roadworks during the summer school, impacting the tram routes. As a result, there won’t be any tram circulating between the stations "Republique" and "Observatoire". Replacement busses will be put into circulation.
You can read the document below for more details.
The map of the central campus is available here.
Photos by Jonathan Martz and Geneviève Engel.
no publication
Crash course in Chemoinformatics
The crash-course will take place on Monday, June 27, from 8:30 a.m. to 4:15 p.m.
Contact : Dr. Gilles Marcou
Interactive Workshop with SeeSAR
Advanced 3D Modeling: LO, FBLD, Ph4 - and More
Marcus Gastreich (BioSolveIT Germany)
Beyond the obvious tasks, SeeSAR can also swiftly be used for more advanced tasks. In this workshop that addresses scenarios where the non-obvious is needed or where roads get bumpy: We shall use the tool to overcome hurdles, realize scenarios when NOT to trust the computer blindly, and hunt for good, well-fitting molecules. This for example refers to:
- My ∆G is far off. What now?
- That docking does not work. How can I proceed?
- I am in desperate need for another core here.
- But I need that interaction with that amino acid up there.
- The boss wants me to come up with fragments attached that explore this sub-pocket.
Using indexing techniques, pharmacophores in combination with SMARTS expressions, and several other functionalities, we will discuss remedies for such scenarios.
If time permits, we will also navigate through chemical spaces of multi-billion member sizes.
Requirements:
You will use your own computer! Bring it along, and make sure you have:
Tutorials of the school
no publication
Tutorial on GTM Landscapes
The tutorial aims at presenting the Generative Topographic Mapping (GTM) landscapes . For more details about the GTM algorithm, see [Bishop et al, Neural Computation 10, No. 1, 215–234 (1998)]. For details about GTM predictive models and landscapes, see [H.A. Gaspar et al. Mol. Informatics, 2015, 34 (6-7), 348-356]
The GTM is an unsupervised method to map high dimensional data to a two-dimensional representation. In the process, the GTM builds a probabilistic model of the data that can be exploited for data characterization, comparison or classification and regression model building. The GTM approach will be used to analyze a bioconcentration factor dataset and to explore structure-activity relationships. As a result, several property and activity landscapes will be generated and visualized.
Software
The tutorial is based on six pieces of software:
- xGTMapTool: a graphical user interface frontend for the preparation of a GTM.
- xGTMview: an application to link the GTM trained on chemical data and the chemical structures.
- xGTMReSample: an application to improve the resolution of a GTM model.
- xGTMReg: an application to build and validate a GTM-based regression model.
- xGTMClass: an application to build and validate a GTM-based classification model.
- xGTMLandscape: an application to visualize and navigate the activity and property landscapes.
The software are supplied online and can be downloaded for the OS of your choice.:
- Windows (Win10, 64bits)
- Mac (for Intel: Monterey 12.4. For M1: BigSur or Monterey 12.4)
- Linux (Ubuntu 20 64bits or Ubuntu 20 64 bits)
Download link (Contains Software + License - valid until the 1st of August, 2022)
Software available until the 17th of July, 2022
Later, you can ask for updated versions here:
http://infochim.u-strasbg.fr/spip.php?rubrique41
The license of the software is distributed freely and a license file, called "licence.dat_CS32022" (to be renamed to "licence.dat") is distributed with the software for the OS of your choice (Windows, Mac or Linux).
The license file must be installed in a proper location to be found.
- On Windows: create the directory AppData\local\ISIDAGTM directory at the root of your home directory and copy the file license.dat in it. The absolute path of the file should be similar to this one:
C:\Users\username\AppData\local\ISIDAGTM\licence.dat
The file and the directory should have read and write permissions.
- On Mac: create the directory .config/ISIDAGTM directory at the root of your home directory and copy the file license.dat in it. The absolute path of the file should be similar to this one:
/Users/username/.config/ISIDAGTM/licence.dat
- On Linux: create the directory .config/ISIDAGTM directory at the root of your home directory and copy the file license.dat in it. The absolute path of the file should be similar to this one:
/home/username/.config/ISIDAGTM/licence.dat
Datasets and Step-by-step instructions
The tutorial uses a dataset of bioconcentration factor Lunghini, F.et al. QSAR & Env. Protection, 30(7), 507-524].
The dataset and instructions can be downloaded here.
Tutorial on protein cavities alignment with ProCare
The tutorial will show you how to use ProCare to align protein cavities. A cavity is described as point clouds in which each point has a pharmacophoric property [J. Desaphy et al. J. Chem. Inf. Model., vol. 52, no 8, p. 2287‑2299 (2012)]. ProCare aligns two point clouds using a Point Cloud Registration algorithm allowing the comparison of cavities of different size (local alignment) [M. Eguida et al. J. Med. Chem., vol. 63, no 13, p. 7127‑7142 (2020)]. In this tutorial, co-crystallized fragmented ligands from the Protein Data Bank will be aligned onto the cavity of a target protein to suggest seed fragments for the de novo construction of a ligand.
Material
The complete tutorial material can be downloaded here:
https://drugdesign.unistra.fr/CS3/2022_
NB: The exercise will be done on your personal computer. To speed up preparation time, please install all necessary software BEFORE the session begins.
Installation
The ProCare software runs only on Linux operating systems. If you use another operating system, it is possible to run ProCare using virtualization software.
Linux
First download the “CS3_2022_procare.zip” archive and unzip it. Then follow the instructions given in the output directory to install software and scripts. Be careful! 7 Gb of free space is required!
MacOS or Windows
You will need the VirtualBox software installed on your computer. You can download this software by clicking on the corresponding link:
• Windows: https://download.virtualbox.org/virtualbox/6.1.34/VirtualBox-6.1.34a-150636-Win.exe
• MacOS: https://download.virtualbox.org/virtualbox/6.1.34/VirtualBox-6.1.34-150636-OSX.dmg
NB1: It is possible to use VMware software for this tutorial if you already installed it.
NB2: For Mac users, VirtualBox must has permission to access the computer. After installation, go to the Security and Privacy panel to change the security rule.
Then download the “Ubuntu_CS3_2022.zip” archive and unzip it. Be careful! 14 Gb of free space is required! The output directory contains the virtual machine file named “Ubuntu_CS3_2022.vmdk” in which there is an Ubuntu distribution with everything already installed.
To login use the following credentials:
• username: puser
• password: 1234